LanGauge Benchmark Blog

PREFACE
Academic research is being published at an astonishing rate. ArXiv has grown exponentially, from 8,871 papers published in October 2015 to 16,418 in October 2020 [1]. With this increase in publications, keeping up with recent advances can be a full-time job.
Previously, we announced the private-alpha release of LanGauge, our biomedical Named-Entity Recognition (NER) computing framework based on Microsoft’s Biomedical Language Understanding and Reasoning Benchmark (BLURB). Named entity recognition means that we can extract key terms and concepts from long papers to save the time of busy medical professionals.
Using LanGauge, bioscientists and pharmaceutical researchers can easily create pipelines for tasks such as NER, reducing the amount of required code for such tasks. LanGauge is open-source, highly extensible, and easy to use via a web-based user interface. To learn more, go to www.langauge.org.
INTRODUCTION
In this blog, we will present our benchmark study in evaluating BLURB on LanGauge. Using 5 datasets encompassing gene/disease mentions, chemical-disease relations, molecular biology concepts were extracted from abstracts of papers using fine-tuned models deployed to LanGauge. We will describe these models and datasets and present our experimental evaluation of their F1 scores on a desktop running a beta version of LanGauge and equipped with an Nvidia GeForce RTX3090. We also present a preliminary analysis of the dataset on OpenAI’s GPT-3.
DATASET
There are 5 constituent datasets of BLURB, all spanning the medical field. They are suited for different tasks and datasets, as follows.
BC2GM
BC2GM [2] is a gene recognition task consisting of sentences from PubMed (MedLine) abstracts were human annotated. Of 20,000 sentences, there are approximately 44,500 gene annotations.
Example:
Input: Physical mapping 220 kb centromeric of the human MHC and DNA sequence analysis of the 43 - kb segment including the RING1, HKE6, and HKE4 genes.
Output:
- human MHC
- RING1
- HKE6
- HKE4 genes
BC5CDR
BC5-CDR [3] is a dataset consisting of chemical and disease relation and named entity extraction. 1500 PubMed (medline) articles with 4409 annotated chemicals, 5818 diseases, and 3116 chemical-disease interactions were used. Each entity annotation includes both the mention text spans and normalized concept identifiers, using MeSH as the controlled vocabulary.
Note that our PubMedBERT was finetuned on BC5CDR-chem and BC5CDR-disease datasets separately.
BC5CDR-chem Example:
Input: The mechanisms of proarrhythmic effects of Dubutamine are discussed.
Output:
- Dubtamine
JNLPBA
JNLPBA [4] consists of PubMed (MedLine) abstracts to classify concepts of interests to biologists in the domain of molecular biology.
Example:
Input: Number of glucocorticoid receptors in lymphocytes and their sensitivity to hormone action
Output:
- glucocorticoid receptors
- lymphocytes
NCBI-disease
The NCBI-disease [5] corpus contains 6,892 disease mentions, which are mapped to 790 unique disease concepts from PubMed (MedLine) abstracts.
Example:
Input: Clustering of missense mutations in the ataxia - telangiectasia gene in a sporadic T - cell leukaemia.
Output:
- ataxia - telangiectasia
- sporadic T-cell leukaemia
MODELS
Our model is a variant of BERT called PubMedBERT by Gu. Y et al. and Microsoft. Each dataset has its own finetuned model. We used PyTorch and Huggingface’s token classification pipeline with largely default hyperparameters:
EVALUATION
Each of the mentioned datasets have pre-split dev, train, and test sets. Our primary metric was entity-level F1 as a result of the unbalanced dataset. When an entity was split into multiple tokens, the first token was used, as in BERT. Through the Jupyter notebook eval.ipynb, it is also possible for the user to input an abstract and a ground truth to evaluate the model on.
The results on these preset test sets are as follows.
Table 1. Execution time on Nvidia GeForce RTX3090.
BC2GM:
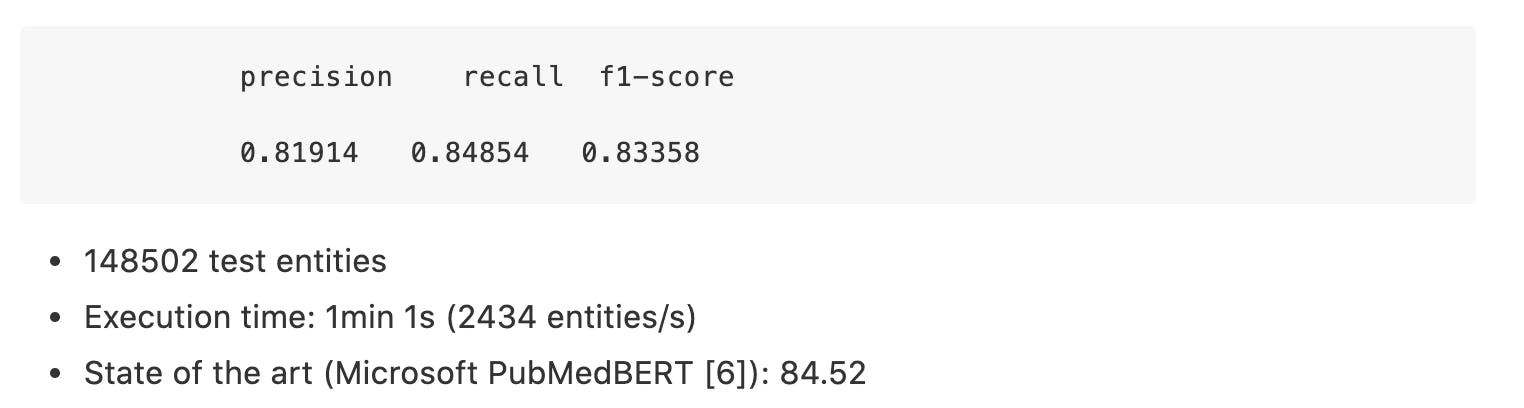
BC5CDR-Chem:
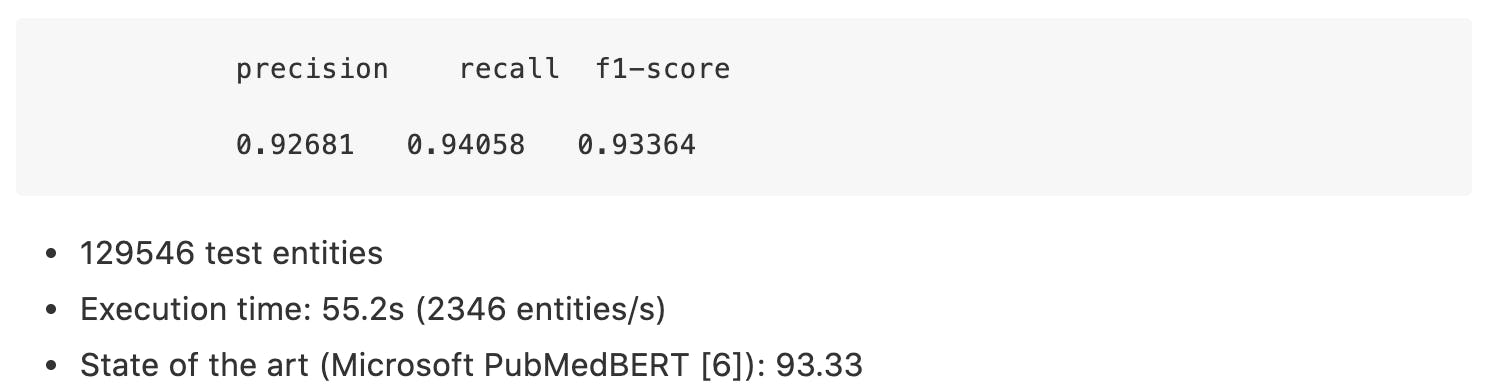
BC5CDR Disease:

JNLPBA:

NCBI Disease:

EXPERIMENTING WITH GPT-3
Considering the recent successes of GPT-3, it may seem like a logical candidate for a task like ours. We obtained a beta API key for GPT-3, and ran some experiments with few-shot NER. GPT-3 is great for few-shot tasks. However, given our large dataset, we also had the capabilities to do many-shot learning, expecting vastly improved results. First, we ran a sanity check to find nouns with only one example.
What are the noun phrases in the following passage?
Passage:
The baseball bat hit the ground as the ball soared through the air. He ran to first base, second, third, and rounded the bend to home.
Nouns:
- baseball bat
- ground
- ball
- air
- first base
- second base
- third base
- bend
- home
It’s perfect, matching the ground truth. Moving on to biomedical NER however, everything started to fall apart. We provided 10 examples to establish a few-shot baseline with GPT-3 on each of our datasets. However, GPT-3 performs abysmally on the test sets, achieving under 0.6 F1 score on each. The below sample is an example on BC2GM:
What are the genes in the following passage?
Physical mapping 220 kb centromeric of the human MHC and DNA sequence analysis of the 43 - kb segment including the RING1, HKE6, and HKE4 genes.
Extracted genes:
Output:
- RING1
- HKE6
- HKE4
- END
The ground truth is:
- human MHC
- RING1
- HKE6
- HKE4 genes
- END
Unfortunately, GPT-3 is a fair ways off; we tried with 50 few-shot examples, and yet results did not improve significantly. Other examples yield similar results, and tuning hyperparameters did not improve performance significantly.
One possible cause for this is the relative rarity of this data, as genes are not commonly seen or associated with each other. This can be remedied by finetuning on these datasets. Since few-shot learning doesn't perform any gradient updates, it’s difficult to use GPT-3 with biomedical NER. OpenAI provides a finetuning library, for python OpenAI -ft, however, we were unable to use it at this time due to an authentication bug, or perhaps only certain users have the privileges to finetune GPT-3. GPT-3 has outstanding performance on everyday language tasks given its training corpus; however, for the time being, it seems that gene recognition is beyond the capabilities of a few shot model.
REFERENCES:
[1] Monthly submissions. (n.d.). Retrieved February 5, 2021.
[2] Smith, L., Tanabe, L. K., Ando, R. J. nee, Kuo, C.-J., Chung, I.-F., Hsu, C.-N., Lin, Y.-S., Klinger, R., Friedrich, C. M., Ganchev, K., Torii, M., Liu, H., Haddow, B., Struble, C. A., Povinelli, R. J., Vlachos, A., Baumgartner, W. A., Hunter, L., Carpenter, B., … Wilbur, W. J. (2008). Overview of BioCreative II gene mention recognition. Genome Biology, 9(2), S2.
[3] Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H., Leaman, R., Davis, A. P., Mattingly, C. J., Wiegers, T. C., & Lu, Z. (2016). BioCreative V CDR task corpus: A resource for chemical disease relation extraction. Database: The Journal of Biological Databases and Curation, 2016.
[4] Collier, N., & Kim, J.-D. (2004). Introduction to the bio-entity recognition task at jnlpba. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications (NLPBA/BioNLP), 73–78.
[5] Doğan, R. I., Leaman, R., & Lu, Z. (2014). Ncbi disease corpus: A resource for disease name recognition and concept normalization. Journal of Biomedical Informatics, 47, 1–10.
[6] Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama, N., Liu, X., Naumann, T., Gao, J., & Poon, H. (2020). Domain-specific language model pretraining for biomedical natural language processing. ArXiv:2007.15779 [Cs].
[7] Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2019). BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, btz682.